Multiple Instance Learning (MIL)
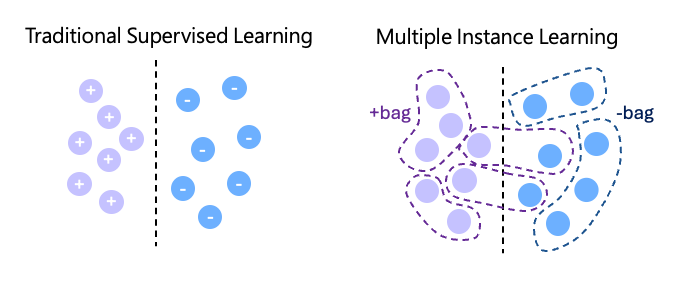
15 minutesMultiple Instance Learning (MIL) is a machine learning approach commonly used for training models on pathology data. In traditional supervised learning, each individual sample is associated with its own label. In contrast, MIL organizes training data into bags, where each bag contains multiple instances, and a single label is assigned to the entire bag rather than to individual instances.
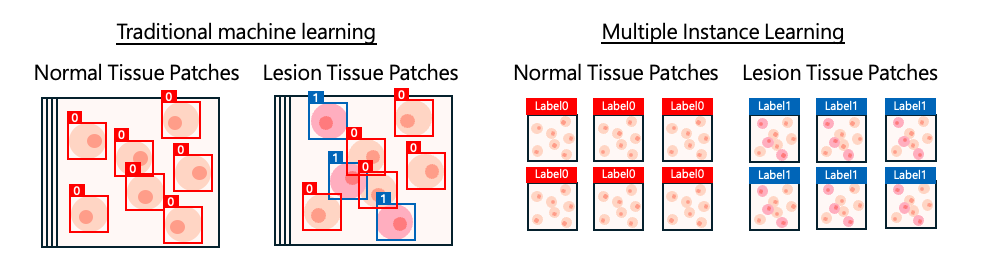
This greatly reduces the need for detailed cell-level annotation—especially useful in tasks such as tumor detection or lesion analysis—because annotators only need to label whole regions or whole-slide images (WSIs) instead of marking every single cell.
The model learns discriminative features from the instances within each bag and produces a final prediction for the bag, such as determining whether the sample contains tumor or abnormal tissue.
Practical Use of Multiple Instance Learning
You may read: CLAM:MIL first.
In the CLAM framework, directly training on the entire WSI is computationally infeasible.
Therefore, during training, the WSI is first divided into a large number of smaller patches, which are then fed into the model.
Manually annotating every single cell or every patch would require enormous effort.
Using MIL, pathologists no longer need to annotate individual cells. Assigning a single label to the whole WSI is sufficient to begin training, greatly reducing the time and workload required on the annotation side.
In addition to saving time, MIL also helps address uncertainty within WSIs.
Because all patches from the same WSI share the same label, the model becomes more robust in recognizing atypical or ambiguous tissue patterns, improving diagnostic accuracy in regions where morphology varies.