Applications of Unsupervised Neural Networks in Cancer Tissue Heterogeneity — The Faust Algorithm
26 minutesWith the rapid advancement of deep learning in recent years, its application in pathology image analysis has become increasingly widespread. However, many existing models still rely heavily on supervised learning, and labeled data in medical imaging is often difficult and labor-intensive to obtain. To address this limitation, the Faust research team proposed an unsupervised model trained on the histological features of RCC (renal cell carcinoma). Their approach greatly expands the applicability of deep learning in computational pathology by removing the dependency on manual annotations.
Training Data Source
The team collected 550 H&E-stained RCC whole-slide images (WSIs) from the publicly available TCGA (The Cancer Genome Atlas) dataset as training data. RCC was chosen because its tumor tissue presents distinct morphological patterns, making it suitable for building an initial unsupervised framework.
Feature Extraction Using the VGG-19 Model
The researchers first applied the VGG-19 model, using ImageNet-pretrained weights, to extract morphological features from WSIs.
By averaging the feature vectors from the final pooling layer, they observed that these vectors captured meaningful histological characteristics such as fibrosis, mucin, and epithelial structures.
Importantly, these features were not dependent on tumor subtype, disease stage, or severity.
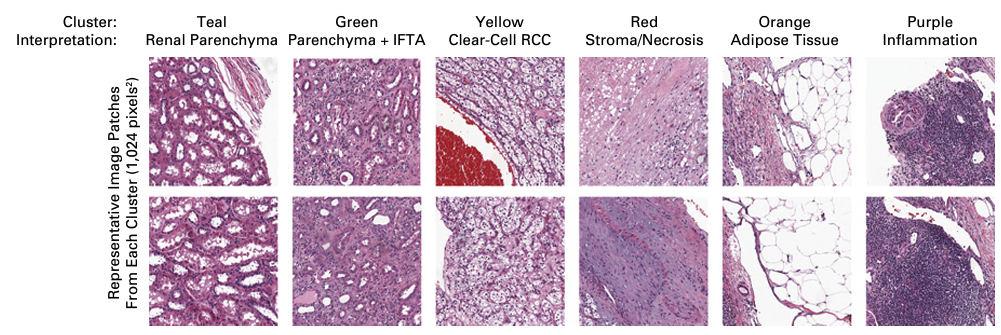
Unsupervised Clustering and Analysis
After obtaining feature vectors from the CNN, the team performed unsupervised clustering. They used the Silhouette Method to determine the optimal number of clusters and applied Ward’s Minimum Variance Method for hierarchical clustering.
In addition to broad-level tissue pattern separation, K-means clustering was used for finer subgrouping, successfully identifying RCC subtypes and correlating clusters with clinical variables such as survival outcomes.
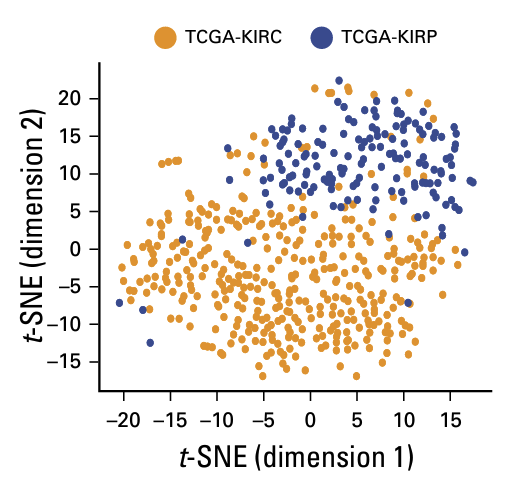
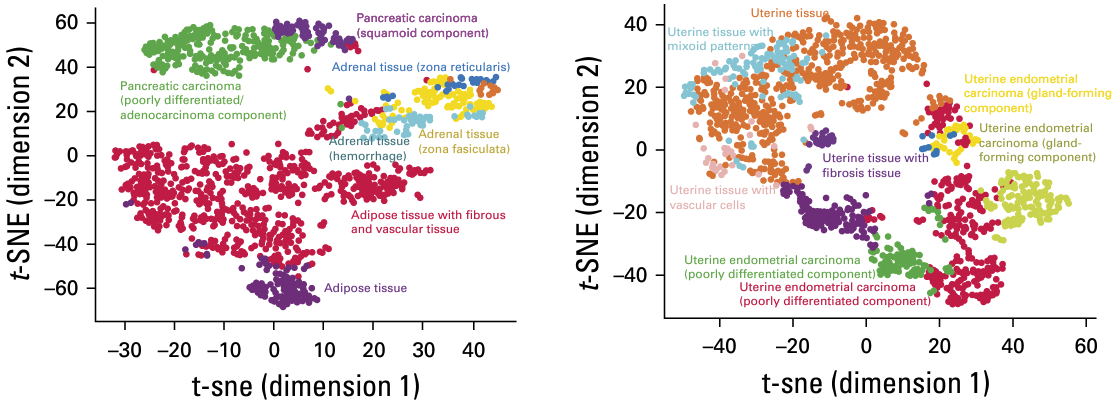
The trained model also demonstrated strong generalizability, effectively clustering tissues from cancer types it had never encountered, such as endometrial carcinoma and pancreatic cancer.
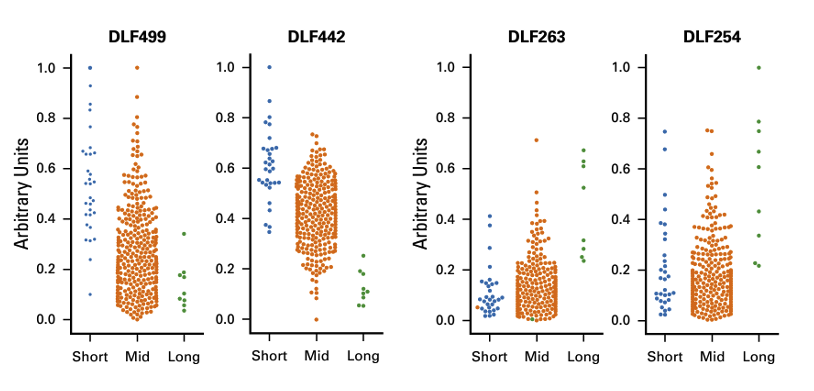
Furthermore, when the clustering results for KIRC were compared to survival data, the team found significant differences in prognosis among clusters.
They identified the DLF (Deep Learned Feature) score as a meaningful indicator correlated with short-, mid-, and long-term survival outcomes.
Conclusion
This study demonstrates that even without manual annotations, unsupervised deep learning can effectively distinguish RCC from other tissues, identify RCC subtypes, and reflect clinically relevant characteristics. Moreover, the framework generalizes well to unseen cancer types, providing a dynamic and objective tool for exploring tumor heterogeneity and offering promising value in prognosis prediction.
Reference
Kevin Faust et al.,Unsupervised Resolution of Histomorphologic Heterogeneity in Renal Cell Carcinoma Using a Brain Tumor–Educated Neural Network. JCO Clin Cancer Inform 4, 811-821(2020).
